Tidying our mean sd function
Write a tidy method for our mean_sd function and try it out on the penguins dataset.
Attaching package: 'palmerpenguins'
The following objects are masked from 'package:datasets':
penguins, penguins_raw
library (broom)#Write a tidy method for our mean_sd function and try it out on the penguins dataset. <- function (x, na.rm = TRUE ) {<- deparse1 (substitute (x)) # gets the name of the variable <- c (mean = mean (x, na.rm = na.rm), sd = sd (x, na.rm = na.rm))class (res) <- "meansd" attr (res, "variable" ) <- xnameattr (res, "sampsize" ) <- length (x)<- function (x, digits = 2 ) {cat (attr (x, "variable" ), ": " ,paste0 (round (x["mean" ], digits = digits), " (" , round (x["sd" ], digits = digits), ") \n " ))<- function (x) {data.frame (variable = attr (x, "variable" ), mean = x["mean" ], sd = x["sd" ], sample.size = attr (x, "sampsize" )mean_sd (penguins$ body_mass_g)
penguins$body_mass_g : 4201.75 (801.95)
tidy (mean_sd (penguins$ body_mass_g))
variable mean sd sample.size
mean penguins$body_mass_g 4201.754 801.9545 344
Apply the mean_sd function to the penguins body mass in grams by species and sex. Organize the results into a table suitable for publication, where it is easy to compare the two sexes.
<- split (penguins$ body_mass_g, list (penguins$ species, penguins$ sex)) |> lapply (FUN = \(x) tidy (mean_sd (x))) <- cbind (do.call (rbind, reslist), do.call (rbind, names (reslist)|> strsplit (split= " \\ ." )))colnames (resdf)[5 : 6 ] <- c ("species" , "sex" )reshape (resdf[, - 1 ], direction = "wide" , idvar = "species" , timevar = "sex" )
species mean.female sd.female sample.size.female mean.male
Adelie.female Adelie 3368.836 269.3801 73 4043.493
Chinstrap.female Chinstrap 3527.206 285.3339 34 3938.971
Gentoo.female Gentoo 4679.741 281.5783 58 5484.836
sd.male sample.size.male
Adelie.female 346.8116 73
Chinstrap.female 362.1376 34
Gentoo.female 313.1586 61
library (data.table)<- data.table (penguins)<- pengdt[, (tidy (mean_sd (body_mass_g))), = list (species, sex)]dcast (peng_summary[! is.na (sex)], species + variable ~ sex, value.var = c ("mean" , "sd" , "sample.size" ))
Key: <species, variable>
species variable mean_female mean_male sd_female sd_male
<fctr> <char> <num> <num> <num> <num>
1: Adelie body_mass_g 3368.836 4043.493 269.3801 346.8116
2: Chinstrap body_mass_g 3527.206 3938.971 285.3339 362.1376
3: Gentoo body_mass_g 4679.741 5484.836 281.5783 313.1586
sample.size_female sample.size_male
<int> <int>
1: 73 73
2: 34 34
3: 58 61
Attaching package: 'dplyr'
The following objects are masked from 'package:data.table':
between, first, last
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
library (tidyr)|> group_by (species, sex) |> summarize (tidy (mean_sd (body_mass_g))) |> filter (! is.na (sex)) |> pivot_wider (names_from = "sex" , values_from = c ("mean" , "sd" , "sample.size" ))
`summarise()` has grouped output by 'species'. You can override using the
`.groups` argument.
# A tibble: 3 × 8
# Groups: species [3]
species variable mean_female mean_male sd_female sd_male sample.size_female
<fct> <chr> <dbl> <dbl> <dbl> <dbl> <int>
1 Adelie body_mas… 3369. 4043. 269. 347. 73
2 Chinstrap body_mas… 3527. 3939. 285. 362. 34
3 Gentoo body_mas… 4680. 5485. 282. 313. 58
# ℹ 1 more variable: sample.size_male <int>
Tidying the national patient register dataset
Load the LPR data example from "https://sachsmc.github.io/r-programming/data/lpr-ex.rds"
here() starts at /home/micsac/Teaching/Courses/r-programming
<- readRDS (here ("data" , "lpr-ex.rds" ))
Reshape the data into wide, where the columns are the primary diagnosis (hdia) at each visit number
Reshape the data into longer format, where all of the diagnoses are stored in a single variable, with another variable indicating the primary diagnosis.
Create a new variable for each participant which equals TRUE if they had any diagnosis of either D150, D152, or D159 before the date 1 January 2010.
## 1. <- subset (lpr, select= c (pid, age, sex, visit, hdia)) |> ## drop the other diag columns and date reshape (direction = "wide" , idvar = "pid" , timevar = "visit" , v.names = c ("hdia" ))head (lprwide)
pid age sex hdia.1 hdia.2 hdia.3 hdia.4 hdia.5 hdia.6 hdia.7 hdia.8 hdia.9
1 A001 72.5 f H560 C871 C040 F412 F622 <NA> <NA> <NA> <NA>
6 A002 81.5 f B481 K959 G902 <NA> <NA> <NA> <NA> <NA> <NA>
9 A003 54.5 m E841 E071 H189 B090 C031 E179 E950 A100 K429
18 A004 72.5 f D921 B401 B491 G172 E161 <NA> <NA> <NA> <NA>
23 A005 86.0 f J751 B101 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
25 A006 77.0 f E310 C231 C212 B251 <NA> <NA> <NA> <NA> <NA>
hdia.10 hdia.11 hdia.12 hdia.13
1 <NA> <NA> <NA> <NA>
6 <NA> <NA> <NA> <NA>
9 <NA> <NA> <NA> <NA>
18 <NA> <NA> <NA> <NA>
23 <NA> <NA> <NA> <NA>
25 <NA> <NA> <NA> <NA>
## 2. <- lprcolnames (lprtmp)[which (colnames (lprtmp) == "hdia" )] <- "diag0" <- reshape (lprtmp, direction = "long" , varying = grep ("^diag" , names (lprtmp)), sep = "" , timevar = "diagnum" )$ maindiag <- lprlong$ diagnum == 0 <- subset (lprlong[order (lprlong$ pid, lprlong$ visit),], ! is.na (diag))head (lprlong)
pid age sex indat visit diagnum diag id maindiag
1.0 A001 72.5 f 2010-01-27 1 0 H560 1 TRUE
1.1 A001 72.5 f 2010-01-27 1 1 B632 1 FALSE
1.2 A001 72.5 f 2010-01-27 1 2 H180 1 FALSE
1.3 A001 72.5 f 2010-01-27 1 3 J050 1 FALSE
2.0 A001 72.5 f 2010-06-26 2 0 C871 2 TRUE
2.1 A001 72.5 f 2010-06-26 2 1 D422 2 FALSE
## 3. <- split (lprlong, lprlong$ pid) |> lapply (FUN = \(df) {<- any (df$ diag[df$ indat <= as.Date ("2010-01-01" )] %in% c ("D150" , "D152" , "D159" ))if (length (chk) == 0 ) chk <- FALSE $ ddiag_pre2010 <- chk|> unsplit (lprlong$ pid)summary (lprlong)
pid age sex indat
Length:8046 Min. :36.50 Length:8046 Min. :2005-01-02
Class :character 1st Qu.:63.50 Class :character 1st Qu.:2007-09-23
Mode :character Median :68.00 Mode :character Median :2010-06-01
Mean :68.43 Mean :2010-06-15
3rd Qu.:77.00 3rd Qu.:2013-03-01
Max. :90.50 Max. :2015-12-31
visit diagnum diag id
Min. : 1.000 Min. :0.000 Length:8046 Min. : 1.0
1st Qu.: 2.000 1st Qu.:1.000 Class :character 1st Qu.: 495.2
Median : 3.000 Median :2.000 Mode :character Median : 997.0
Mean : 3.466 Mean :1.791 Mean : 996.5
3rd Qu.: 5.000 3rd Qu.:3.000 3rd Qu.:1494.0
Max. :13.000 Max. :6.000 Max. :2008.0
maindiag ddiag_pre2010
Mode :logical Mode :logical
FALSE:6038 FALSE:8032
TRUE :2008 TRUE :14
## 1. <- data.table (lpr)dcast (lprdt[, .(pid, indat, visit, hdia)], ~ visit, value.var = "hdia" )
Key: <pid>
pid 1 2 3 4 5 6 7 8 9
<char> <char> <char> <char> <char> <char> <char> <char> <char> <char>
1: A001 H560 C871 C040 F412 F622 <NA> <NA> <NA> <NA>
2: A002 B481 K959 G902 <NA> <NA> <NA> <NA> <NA> <NA>
3: A003 E841 E071 H189 B090 C031 E179 E950 A100 K429
4: A004 D921 B401 B491 G172 E161 <NA> <NA> <NA> <NA>
5: A005 J751 B101 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
---
396: A396 G791 G580 A291 E282 <NA> <NA> <NA> <NA> <NA>
397: A397 K931 K922 G219 J462 I250 <NA> <NA> <NA> <NA>
398: A398 D551 C280 F540 K249 D360 G181 I889 <NA> <NA>
399: A399 E761 E262 G000 <NA> <NA> <NA> <NA> <NA> <NA>
400: A400 I291 E461 F581 D282 J149 <NA> <NA> <NA> <NA>
10 11 12 13
<char> <char> <char> <char>
1: <NA> <NA> <NA> <NA>
2: <NA> <NA> <NA> <NA>
3: <NA> <NA> <NA> <NA>
4: <NA> <NA> <NA> <NA>
5: <NA> <NA> <NA> <NA>
---
396: <NA> <NA> <NA> <NA>
397: <NA> <NA> <NA> <NA>
398: <NA> <NA> <NA> <NA>
399: <NA> <NA> <NA> <NA>
400: <NA> <NA> <NA> <NA>
# 2. <- melt (lprdt, id.vars = c ("pid" , "age" , "sex" , "indat" , "visit" ), variable.name = "diagtype" , value.name = "diag" )<- lprlong[! is.na (diag)]: = diagtype == "hdia" ]
pid age sex indat visit diagtype diag main_diag
<char> <num> <char> <Date> <int> <fctr> <char> <lgcl>
1: A001 72.5 f 2010-01-27 1 hdia H560 TRUE
2: A001 72.5 f 2010-06-26 2 hdia C871 TRUE
3: A001 72.5 f 2010-07-20 3 hdia C040 TRUE
4: A001 72.5 f 2011-03-06 4 hdia F412 TRUE
5: A001 72.5 f 2011-12-23 5 hdia F622 TRUE
---
8042: A390 50.0 m 2014-05-29 8 diag6 B329 FALSE
8043: A390 50.0 m 2014-12-31 9 diag6 H441 FALSE
8044: A391 50.0 m 2008-11-26 4 diag6 H381 FALSE
8045: A393 63.5 m 2015-06-08 7 diag6 I372 FALSE
8046: A398 63.5 m 2009-09-12 3 diag6 J671 FALSE
setorder (lprlong, pid, indat)
pid age sex indat visit diagtype diag main_diag
<char> <num> <char> <Date> <int> <fctr> <char> <lgcl>
1: A001 72.5 f 2010-01-27 1 hdia H560 TRUE
2: A001 72.5 f 2010-01-27 1 diag1 B632 FALSE
3: A001 72.5 f 2010-01-27 1 diag2 H180 FALSE
4: A001 72.5 f 2010-01-27 1 diag3 J050 FALSE
5: A001 72.5 f 2010-06-26 2 hdia C871 TRUE
---
8042: A400 72.5 f 2011-10-26 4 diag2 C010 FALSE
8043: A400 72.5 f 2013-09-01 5 hdia J149 TRUE
8044: A400 72.5 f 2013-09-01 5 diag1 A920 FALSE
8045: A400 72.5 f 2013-09-01 5 diag2 F452 FALSE
8046: A400 72.5 f 2013-09-01 5 diag3 K301 FALSE
# 3. <- lprlong[, ddiag_pre2010 : = any (diag[indat <= as.Date ("2010-01-01" )] %in% c ("D150" , "D152" , "D159" )), = .(pid)]summary (lprlong)
pid age sex indat
Length:8046 Min. :36.50 Length:8046 Min. :2005-01-02
Class :character 1st Qu.:63.50 Class :character 1st Qu.:2007-09-23
Mode :character Median :68.00 Mode :character Median :2010-06-01
Mean :68.43 Mean :2010-06-15
3rd Qu.:77.00 3rd Qu.:2013-03-01
Max. :90.50 Max. :2015-12-31
visit diagtype diag main_diag
Min. : 1.000 hdia :2008 Length:8046 Mode :logical
1st Qu.: 2.000 diag1:2008 Class :character FALSE:6038
Median : 3.000 diag2:1629 Mode :character TRUE :2008
Mean : 3.466 diag3:1178
3rd Qu.: 5.000 diag4: 695
Max. :13.000 diag5: 341
diag6: 187
ddiag_pre2010
Mode :logical
FALSE:8032
TRUE :14
pid age sex indat visit hdia diag1 diag2 diag3 diag4
<char> <num> <char> <Date> <int> <char> <char> <char> <char> <char>
1: A048 86 f 2006-08-20 1 I162 E439 D152 F851 C230
2: A048 86 f 2008-03-30 2 A981 D069 C472 E720 G119
diag5 diag6
<char> <char>
1: E249 I801
2: B912 K901
<- as_tibble (lpr)## 1. |> select (pid, age, sex, visit, hdia) |> pivot_wider (names_from = visit, values_from = hdia, names_prefix = "visit" )
# A tibble: 400 × 16
pid age sex visit1 visit2 visit3 visit4 visit5 visit6 visit7 visit8
<chr> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 A001 72.5 f H560 C871 C040 F412 F622 <NA> <NA> <NA>
2 A002 81.5 f B481 K959 G902 <NA> <NA> <NA> <NA> <NA>
3 A003 54.5 m E841 E071 H189 B090 C031 E179 E950 A100
4 A004 72.5 f D921 B401 B491 G172 E161 <NA> <NA> <NA>
5 A005 86 f J751 B101 <NA> <NA> <NA> <NA> <NA> <NA>
6 A006 77 f E310 C231 C212 B251 <NA> <NA> <NA> <NA>
7 A007 86 f I462 F190 <NA> <NA> <NA> <NA> <NA> <NA>
8 A008 90.5 f C451 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
9 A009 72.5 f D162 H580 C729 B992 B500 <NA> <NA> <NA>
10 A010 63.5 m A709 A772 B800 F212 K402 F712 K180 <NA>
# ℹ 390 more rows
# ℹ 5 more variables: visit9 <chr>, visit10 <chr>, visit11 <chr>,
# visit12 <chr>, visit13 <chr>
## 2. <- lprtbl |> pivot_longer (cols = hdia: diag6, values_to = "diag" ) |> mutate (maindiag = name == "hdia" ) |> filter (! is.na (diag))
# A tibble: 8,046 × 8
pid age sex indat visit name diag maindiag
<chr> <dbl> <chr> <date> <int> <chr> <chr> <lgl>
1 A001 72.5 f 2010-01-27 1 hdia H560 TRUE
2 A001 72.5 f 2010-01-27 1 diag1 B632 FALSE
3 A001 72.5 f 2010-01-27 1 diag2 H180 FALSE
4 A001 72.5 f 2010-01-27 1 diag3 J050 FALSE
5 A001 72.5 f 2010-06-26 2 hdia C871 TRUE
6 A001 72.5 f 2010-06-26 2 diag1 D422 FALSE
7 A001 72.5 f 2010-06-26 2 diag2 K820 FALSE
8 A001 72.5 f 2010-06-26 2 diag3 A602 FALSE
9 A001 72.5 f 2010-07-20 3 hdia C040 TRUE
10 A001 72.5 f 2010-07-20 3 diag1 B710 FALSE
# ℹ 8,036 more rows
## 3. <- lprlong |> group_by (pid) |> mutate (ddiag_pre2010 = any (diag[indat <= as.Date ("2010-01-01" )] %in% c ("D150" , "D152" , "D159" )))summary (lprlong)
pid age sex indat
Length:8046 Min. :36.50 Length:8046 Min. :2005-01-02
Class :character 1st Qu.:63.50 Class :character 1st Qu.:2007-09-23
Mode :character Median :68.00 Mode :character Median :2010-06-01
Mean :68.43 Mean :2010-06-15
3rd Qu.:77.00 3rd Qu.:2013-03-01
Max. :90.50 Max. :2015-12-31
visit name diag maindiag
Min. : 1.000 Length:8046 Length:8046 Mode :logical
1st Qu.: 2.000 Class :character Class :character FALSE:6038
Median : 3.000 Mode :character Mode :character TRUE :2008
Mean : 3.466
3rd Qu.: 5.000
Max. :13.000
ddiag_pre2010
Mode :logical
FALSE:8032
TRUE :14
Merging and manipulation
The objective of this exercise is to describe the distribution of the number of days between hospitalizations and drug dispensations by age and sex, among people who get drugs dispensed following a hospitalization.
Import and merge the drug register data with the long version of the hospitalization register from the previous section. What type of join is needed here?
Create a new variable that counts the number of drug dispensations in the 3 months following a hospitalization.
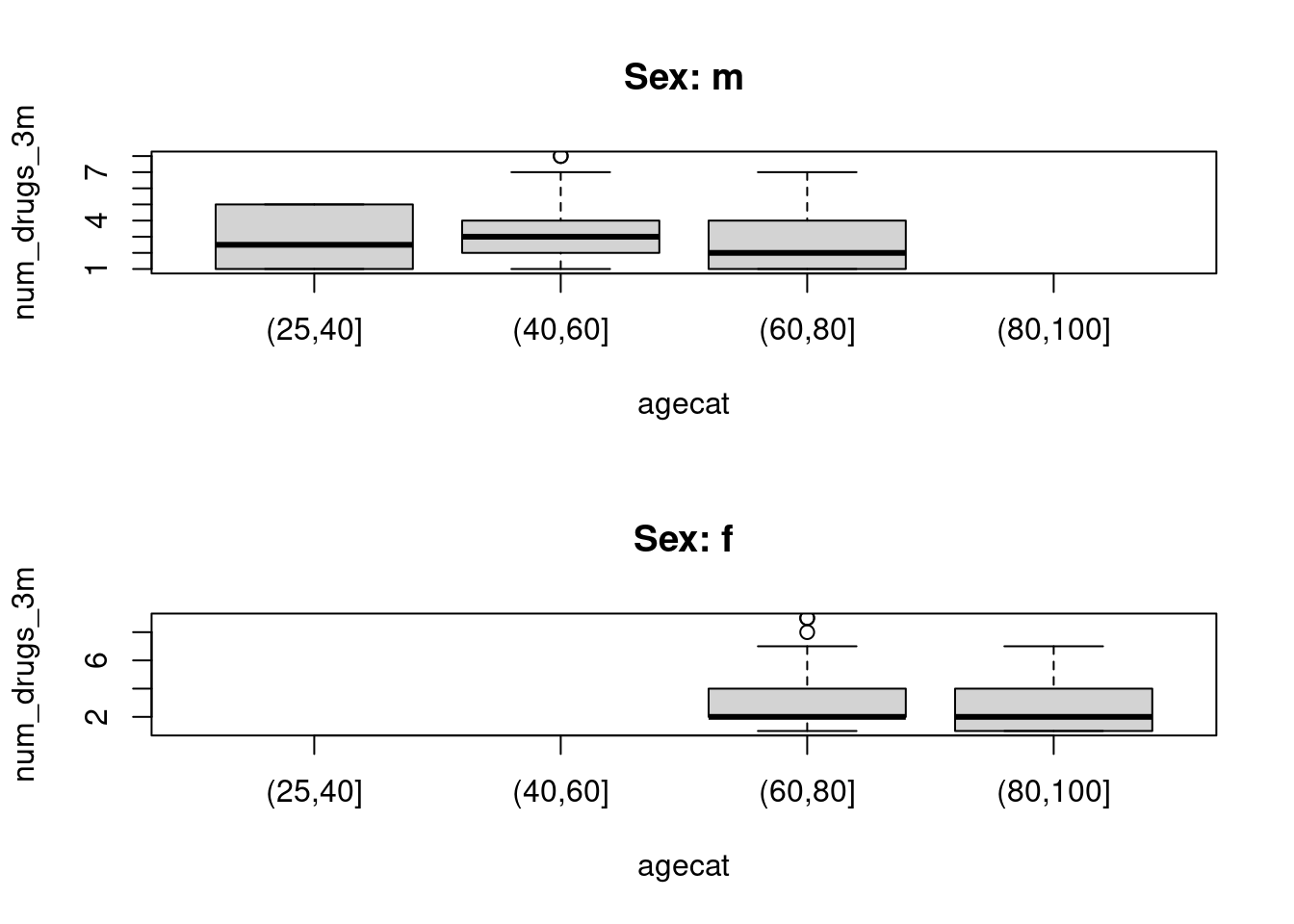
Summarize the variable by age and sex. Try making a graphical summary.
Reading in the data using lapply
<- lapply (2005 : 2010 , \(y) {readRDS (here ("data" , paste0 ("med-" , y, "-ex.rds" )))
or a loop
<- vector (mode = "list" , length = 6 )names (medsl) <- 2005 : 2010 for (i in 1 : length (medsl)) {<- readRDS (here ("data" , paste0 ("med-" , names (medsl)[i], "-ex.rds" )))<- do.call (rbind, medsl)
Attaching package: 'lubridate'
The following objects are masked from 'package:data.table':
hour, isoweek, mday, minute, month, quarter, second, wday, week,
yday, year
The following objects are masked from 'package:base':
date, intersect, setdiff, union
<- merge (subset (lprlong, maindiag == TRUE ), meds, by = "pid" )$ days_to_dispensation <- with (lprmed, date - indat)<- subset (lprmed, days_to_dispensation >= 0 & <= dmonths (3 ))<- do.call ("rbind" , split (lprmed, list (lprmed$ pid, lprmed$ visit), drop = TRUE ) |> lapply (\(df) {$ num_drugs_3m <- nrow (df)unique (df[, c ("pid" , "age" , "sex" , "indat" , "visit" , "diag" , "num_drugs_3m" )])<- lprmed2[order (lprmed2$ pid, lprmed2$ visit),]$ agecat <- cut (lprmed2$ age, c (25 , 40 , 60 , 80 , 100 ))by (lprmed2$ num_drugs_3m, list (lprmed2$ sex, $ agecat),
: f
: (25,40]
NULL
------------------------------------------------------------
: m
: (25,40]
1 2 3 5
2 1 1 2
------------------------------------------------------------
: f
: (40,60]
NULL
------------------------------------------------------------
: m
: (40,60]
1 2 3 4 5 6 7 8
36 56 63 29 17 3 3 2
------------------------------------------------------------
: f
: (60,80]
1 2 3 4 5 6 7 8 9
131 143 116 86 28 14 4 1 2
------------------------------------------------------------
: m
: (60,80]
1 2 3 4 5 6 7
49 46 35 25 12 7 3
------------------------------------------------------------
: f
: (80,100]
1 2 3 4 5 6 7
32 24 24 19 7 1 2
------------------------------------------------------------
: m
: (80,100]
NULL
par (mfrow = c (2 , 1 )) for (s in c ("m" , "f" )) {boxplot (num_drugs_3m ~ agecat, data = subset (lprmed2, sex == s), main = paste0 ("Sex: " , s))
<- lprlong |> filter (maindiag == TRUE ) |> mutate (indat_plus_3m = indat + dmonths (3 )) |> inner_join (meds, join_by (pid, between (y$ date, x$ indat, x$ indat_plus_3m))) |> group_by (pid, visit) |> summarize (sex = sex[1 ], age = age[1 ], agecat = cut (age[1 ], c (25 , 40 , 60 , 80 , 100 )),n_drugs_3m = n ())
`summarise()` has grouped output by 'pid'. You can override using the `.groups`
argument.
|> group_by (sex, agecat, n_drugs_3m) |> summarize (count_ndrugs = n ())
`summarise()` has grouped output by 'sex', 'agecat'. You can override using the
`.groups` argument.
# A tibble: 35 × 4
# Groups: sex, agecat [5]
sex agecat n_drugs_3m count_ndrugs
<chr> <fct> <int> <int>
1 f (60,80] 1 131
2 f (60,80] 2 143
3 f (60,80] 3 116
4 f (60,80] 4 86
5 f (60,80] 5 28
6 f (60,80] 6 14
7 f (60,80] 7 4
8 f (60,80] 8 1
9 f (60,80] 9 2
10 f (80,100] 1 32
# ℹ 25 more rows
with (lprmeddply, by (n_drugs_3m, list (sex, agecat), table))
: f
: (25,40]
NULL
------------------------------------------------------------
: m
: (25,40]
1 2 3 5
2 1 1 2
------------------------------------------------------------
: f
: (40,60]
NULL
------------------------------------------------------------
: m
: (40,60]
1 2 3 4 5 6 7 8
36 56 63 29 17 3 3 2
------------------------------------------------------------
: f
: (60,80]
1 2 3 4 5 6 7 8 9
131 143 116 86 28 14 4 1 2
------------------------------------------------------------
: m
: (60,80]
1 2 3 4 5 6 7
49 46 35 25 12 7 3
------------------------------------------------------------
: f
: (80,100]
1 2 3 4 5 6 7
32 24 24 19 7 1 2
------------------------------------------------------------
: m
: (80,100]
NULL
<- data.table (lprlong)[maindiag == TRUE ]<- data.table (meds): = date]: = indat + dmonths (3 )]<- meds[lprdt, , on = .(pid == pid, date >= indat), nomatch = NULL ][drugdate <= indate_plus_3m]<- lprmeds[, .(sex = sex[1 ], age = age[1 ], agecat = cut (age[1 ], c (25 , 40 , 60 , 80 , 100 )), n_drugs_3m = .N), by = .(pid, visit)]with (lprmeds2, by (n_drugs_3m, list (sex, agecat), table))
: f
: (25,40]
NULL
------------------------------------------------------------
: m
: (25,40]
1 2 3 5
2 1 1 2
------------------------------------------------------------
: f
: (40,60]
NULL
------------------------------------------------------------
: m
: (40,60]
1 2 3 4 5 6 7 8
36 56 63 29 17 3 3 2
------------------------------------------------------------
: f
: (60,80]
1 2 3 4 5 6 7 8 9
131 143 116 86 28 14 4 1 2
------------------------------------------------------------
: m
: (60,80]
1 2 3 4 5 6 7
49 46 35 25 12 7 3
------------------------------------------------------------
: f
: (80,100]
1 2 3 4 5 6 7
32 24 24 19 7 1 2
------------------------------------------------------------
: m
: (80,100]
NULL