Flow control and loops
Flow control
What?
Normally the code gets run line by line, in order, top to bottom
There are special commands that allow you change that
Conditional execution, choose which code to run depending on logical conditions
if(<condition>)else ifelse
Loops, repeat a chunk of code several times
repeatforwhile
If then else
If the <condition> evaluates to TRUE, then something gets executed, otherwise something else gets executed.
<condition> must be length one. No vectors allowed.
Examples
This is more useful when the condition inside the if is a result of some other computation
Examples
Examples
There does not need to be anything returned from the expression, it could just modify data, make a figure, etc.
Many if else
There is no limit to the amount of if else statements. This is equivalent to having if statements repeatedly nested.
Related functions
switch: evaluates its first argument, and chooses one of the subsequent arguments by name (unnamed the “default”)
cut: Divides a numeric vector into intervals and produces a factor.
Logical operators
& (logical AND) and | (OR) have scalar counterparts, called “short-circuit” operators
&&and||evaluate the first argument, and then the second one only if necessary.
Common use case:
Other logical operators
xorExclusive ORisTRUEandisFALSEanyandallfor converting logical vectors to a single value, also havena.rmoption
Exception handling
aka, how to not stop evaluation on an error. R has 3 types of exceptions: 1) Errors, 2) Warnings, 3) Messages. Errors stop the flow of execution, all three say something at the console.
Warnings and Messages can be suppressed by wrapping the expressions in suppress(Warhings|Messages)
tryCatch evaluates an expression and does something with the error that you decide.
tryCatch({ # block of expressions to execute, until an error occurs
cat("a...\n")
stop("b!") # error – breaks the linear control flow
cat("c?\n")
},
error = function(e) { # executed immediately on an error
cat(sprintf("[error] %s\n", e[["message"]]))
},
finally = { # always executed at the end, regardless of error occurrence
cat("d.\n")
}
)a...
[error] b!
d.ifelse
This is a function, not a statement like if and else.
It is vectorized and hence better suited for working with data.
ifelse(<logical vector>, <yes vector>, <no vector>). All three vectors should be the same length or recycling happens.
It returns a vector with elements from yes when TRUE, and elements from no when FALSE.
Loops
Basic concepts
A loop repeatedly and sequentially evaluates an expression, i.e.,
A loop will continue forever unless you tell it to stop. You tell it to stop in different ways for different loop expressions
Repeat
This is the simplest of loops, it will repeat an expression until it encounters break
This will run forever
This will run exactly once
Modify the above to run 5 times
While
Notice the pattern, repeat an expression until a condition is met.
The condition usually depends on a variable that changes at each iteration, in this case i, the iterator
while loops explicitly state the condition at the start:
For loops
A for loop explicitly states the sequence of the iterator at the start. Then the “end condition” is that the loop has reached the end of the sequence.
In this case, it is the most concise. I personally use for loops more than any other loop.
break can be used inside any loop to end it. next can be used to go to the next iteration.
Example - Iterating through species
library(palmerpenguins)
species_names <- levels(penguins$species)
mean_bill_length <- numeric(length(species_names))
names(mean_bill_length) <- species_names
for(i in species_names){ ## iterator is a character
mean_bill_length[i] <- ## indexing by name
mean(subset(penguins, species == i)$bill_length_mm, na.rm = TRUE)
}
mean_bill_length Adelie Chinstrap Gentoo
38.79139 48.83382 47.50488 Bootstrap
mu.body_mass <- mean(penguins$body_mass_g, na.rm = TRUE)
bsmeans <- vector("numeric", length = 200)
for(i in 1:length(bsmeans)) {
resampled.body_mass <- sample(penguins$body_mass_g, replace = TRUE)
bsmeans[i] <- mean(resampled.body_mass, na.rm = TRUE)
}
hist(bsmeans)
abline(v = mu.body_mass, col = "red")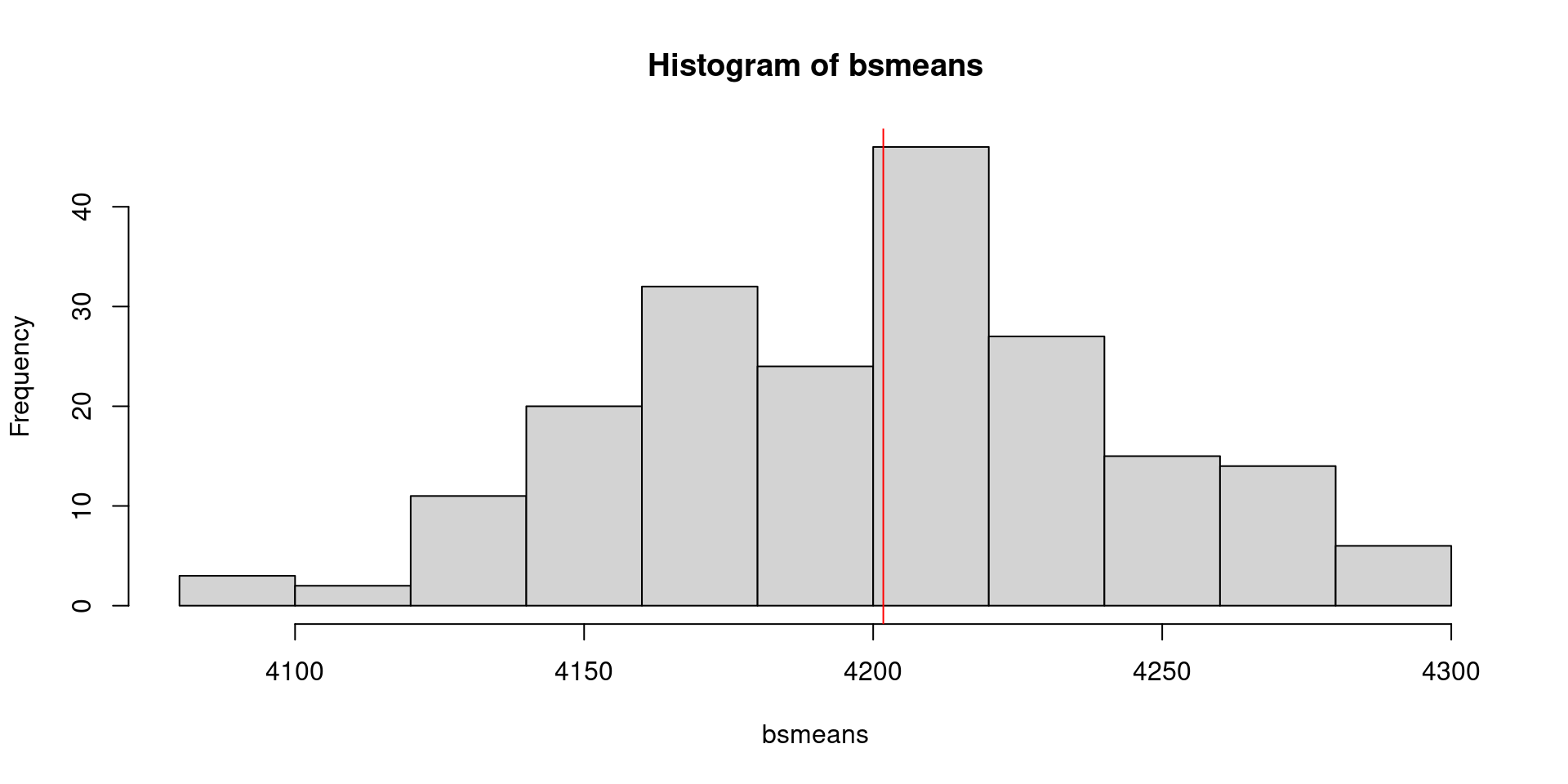
Nested loops
A loop can itself contain a loop, or multiple loops.
species_names <- levels(penguins$species)
island_names <- levels(penguins$island)
mean_bm_matrix <- matrix(NA, nrow = length(species_names),
ncol = length(island_names),
dimnames = list(species_names, island_names))
for(i in species_names) {
for(j in island_names) {
thisset <- subset(penguins, species == i &
island == j)
if(nrow(thisset) == 0) next
mean_bm_matrix[i, j] <- mean(thisset$body_mass_g, na.rm = TRUE)
}
}
mean_bm_matrix Biscoe Dream Torgersen
Adelie 3709.659 3688.393 3706.373
Chinstrap NA 3733.088 NA
Gentoo 5076.016 NA NANote on speed
You may see people warn you not to use for loops in R, “because they are slow”.
That is partially true, but speed is not the only thing, for loops can be much clearer and more understandable than the alternatives.
But, the slow thing in R is changing the size of an object, so you can avoid that by creating a vector/matrix/array of the correct size to hold the results of the loop: