Regression standardization in conditional generalized estimating equations
Source:R/gee_methods.R
standardize_gee.Rdstandardize_gee performs regression standardization in linear and log-linear
fixed effects models, at specified values of the exposure, over the sample
covariate distribution. Let \(Y\), \(X\), and \(Z\) be the outcome,
the exposure, and a vector of covariates, respectively. It is assumed that
data are clustered with a cluster indicator \(i\). standardize_gee uses
fitted fixed effects model, with cluster-specific intercept \(a_i\) (see
details), to estimate the standardized mean
\(\theta(x)=E\{E(Y|i,X=x,Z)\}\), where \(x\) is a specific value of
\(X\), and the outer expectation is over the marginal distribution of
\((a_i,Z)\).
Usage
standardize_gee(
formula,
link = "identity",
data,
values,
clusterid,
case_control = FALSE,
ci_level = 0.95,
ci_type = "plain",
contrasts = NULL,
family = "gaussian",
reference = NULL,
transforms = NULL
)Arguments
- formula
A formula to be used with
"gee"in the drgee package.- link
The link function to be used with
"gee".- data
The data.
- values
A named list or data.frame specifying the variables and values at which marginal means of the outcome will be estimated.
- clusterid
An optional string containing the name of a cluster identification variable when data are clustered.
- case_control
Whether the data comes from a case-control study.
- ci_level
Coverage probability of confidence intervals.
- ci_type
A string, indicating the type of confidence intervals. Either "plain", which gives untransformed intervals, or "log", which gives log-transformed intervals.
- contrasts
A vector of contrasts in the following format: If set to
"difference"or"ratio", then \(\psi(x)-\psi(x_0)\) or \(\psi(x) / \psi(x_0)\) are constructed, where \(x_0\) is a reference level specified by thereferenceargument. Has to beNULLif no references are specified.- family
The family argument which is used to fit the glm model for the outcome.
- reference
A vector of reference levels in the following format: If
contrastsis notNULL, the desired reference level(s). This must be a vector or list the same length ascontrasts, and if not named, it is assumed that the order is as specified in contrasts.- transforms
A vector of transforms in the following format: If set to
"log","logit", or"odds", the standardized mean \(\theta(x)\) is transformed into \(\psi(x)=\log\{\theta(x)\}\), \(\psi(x)=\log[\theta(x)/\{1-\theta(x)\}]\), or \(\psi(x)=\theta(x)/\{1-\theta(x)\}\), respectively. If the vector isNULL, then \(\psi(x)=\theta(x)\).
Value
An object of class std_glm. Obtain numeric results in a data frame with the tidy.std_glm function.
This is a list with the following components:
- res_contrast
An unnamed list with one element for each of the requested contrasts. Each element is itself a list with the elements:
- estimates
Estimated counterfactual means and standard errors for each exposure level
- covariance
Estimated covariance matrix of counterfactual means
- fit_outcome
The estimated regression model for the outcome
- fit_exposure
The estimated exposure model
- exposure_names
A character vector of the exposure variable names
- est_table
Data.frame of the estimates of the contrast with inference
- transform
The transform argument used for this contrast
- contrast
The requested contrast type
- reference
The reference level of the exposure
- ci_type
Confidence interval type
- ci_level
Confidence interval level
- res
A named list with the elements:
- estimates
Estimated counterfactual means and standard errors for each exposure level
- covariance
Estimated covariance matrix of counterfactual means
- fit_outcome
The estimated regression model for the outcome
- fit_exposure
The estimated exposure model
- exposure_names
A character vector of the exposure variable names
Details
standardize_gee assumes that a fixed effects model
$$\eta\{E(Y|i,X,Z)\}=a_i+h(X,Z;\beta)$$ has been fitted. The link
function \(\eta\) is assumed to be the identity link or the log link. The
conditional generalized estimating equation (CGEE) estimate of \(\beta\)
is used to obtain estimates of the cluster-specific means:
$$\hat{a}_i=\sum_{j=1}^{n_i}r_{ij}/n_i,$$ where
$$r_{ij}=Y_{ij}-h(X_{ij},Z_{ij};\hat{\beta})$$ if \(\eta\) is the
identity link, and $$r_{ij}=Y_{ij}\exp\{-h(X_{ij},Z_{ij};\hat{\beta})\}$$
if \(\eta\) is the log link, and \((X_{ij},Z_{ij})\) is the value of
\((X,Z)\) for subject \(j\) in cluster \(i\), \(j=1,...,n_i\),
\(i=1,...,n\). The CGEE estimate of \(\beta\) and the estimate of
\(a_i\) are used to estimate the mean \(E(Y|i,X=x,Z)\):
$$\hat{E}(Y|i,X=x,Z)=\eta^{-1}\{\hat{a}_i+h(X=x,Z;\hat{\beta})\}.$$ For
each \(x\) in the x argument, these estimates are averaged across
all subjects (i.e. all observed values of \(Z\) and all estimated values
of \(a_i\)) to produce estimates $$\hat{\theta}(x)=\sum_{i=1}^n
\sum_{j=1}^{n_i} \hat{E}(Y|i,X=x,Z_i)/N,$$ where \(N=\sum_{i=1}^n n_i\).
The variance for \(\hat{\theta}(x)\) is obtained by the sandwich formula.
Note
The variance calculation performed by standardize_gee does not condition
on the observed covariates \(\bar{Z}=(Z_{11},...,Z_{nn_i})\). To see how
this matters, note that
$$var\{\hat{\theta}(x)\}=E[var\{\hat{\theta}(x)|\bar{Z}\}]+var[E\{\hat{\theta}(x)|\bar{Z}\}].$$
The usual parameter \(\beta\) in a generalized linear model does not
depend on \(\bar{Z}\). Thus, \(E(\hat{\beta}|\bar{Z})\) is independent
of \(\bar{Z}\) as well (since \(E(\hat{\beta}|\bar{Z})=\beta\)), so that
the term \(var[E\{\hat{\beta}|\bar{Z}\}]\) in the corresponding variance
decomposition for \(var(\hat{\beta})\) becomes equal to 0. However,
\(\theta(x)\) depends on \(\bar{Z}\) through the average over the sample
distribution for \(Z\), and thus the term
\(var[E\{\hat{\theta}(x)|\bar{Z}\}]\) is not 0, unless one conditions on
\(\bar{Z}\).
References
Goetgeluk S. and Vansteelandt S. (2008). Conditional generalized estimating equations for the analysis of clustered and longitudinal data. Biometrics 64(3), 772-780.
Martin R.S. (2017). Estimation of average marginal effects in multiplicative unobserved effects panel models. Economics Letters 160, 16-19.
Sjölander A. (2019). Estimation of marginal causal effects in the presence of confounding by cluster. Biostatistics doi: 10.1093/biostatistics/kxz054
Examples
require(drgee)
#> Loading required package: drgee
set.seed(4)
n <- 300
ni <- 2
id <- rep(1:n, each = ni)
ai <- rep(rnorm(n), each = ni)
Z <- rnorm(n * ni)
X <- rnorm(n * ni, mean = ai + Z)
Y <- rnorm(n * ni, mean = ai + X + Z + 0.1 * X^2)
dd <- data.frame(id, Z, X, Y)
fit.std <- standardize_gee(
formula = Y ~ X + Z + I(X^2),
link = "identity",
data = dd,
values = list(X = seq(-3, 3, 0.5)),
clusterid = "id"
)
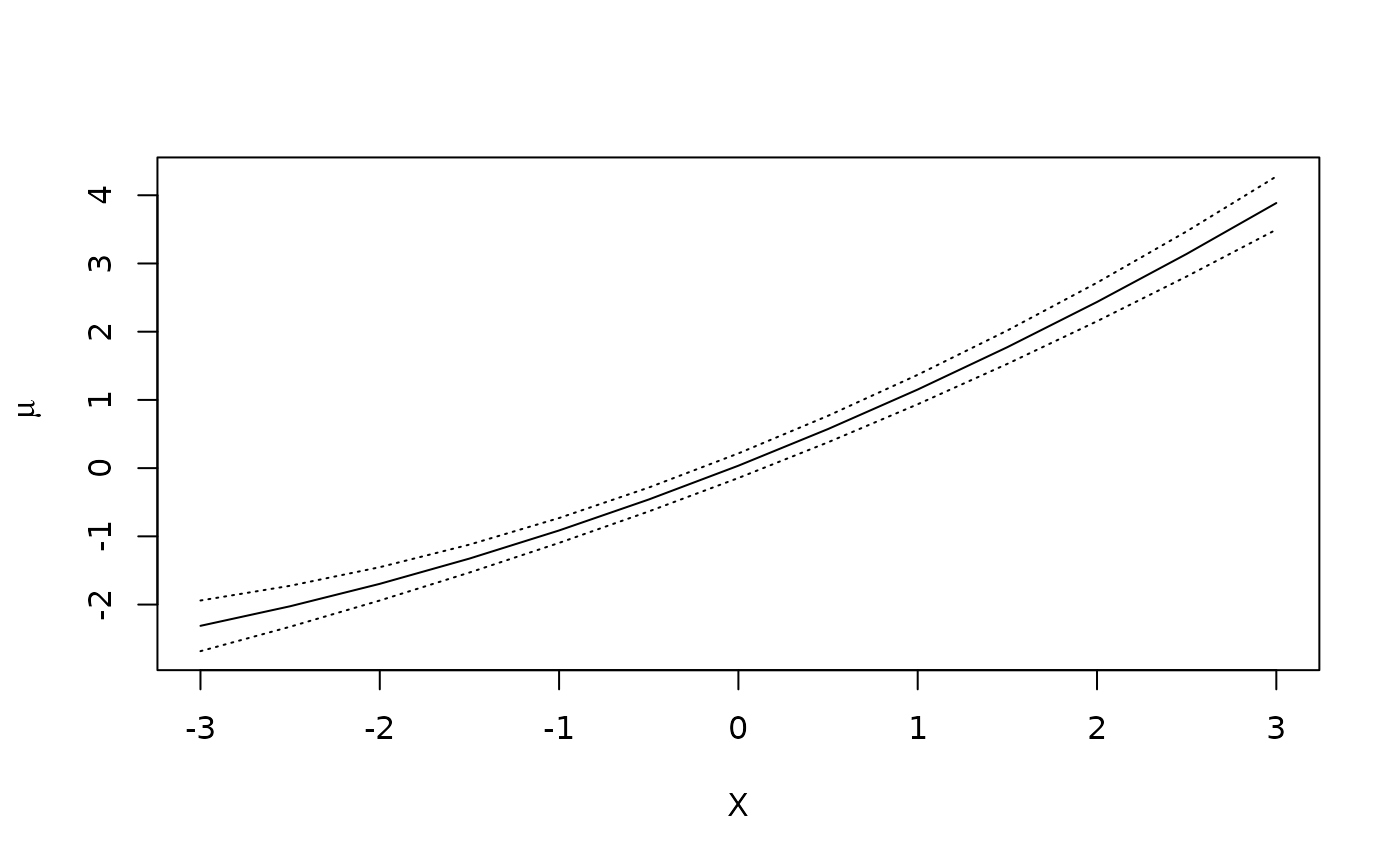
print(fit.std)
#> Outcome formula: Y ~ X + Z + I(X^2)
#> <environment: 0x58fe728f4068>
#> Outcome family:
#> Outcome link function:
#> Exposure: X
#>
#> Tables:
#> X Estimate Std.Error lower.0.95 upper.0.95
#> 1 -3.0 -2.3117 0.1895 -2.683 -1.940
#> 2 -2.5 -2.0248 0.1531 -2.325 -1.725
#> 3 -2.0 -1.6962 0.1246 -1.940 -1.452
#> 4 -1.5 -1.3258 0.1045 -1.531 -1.121
#> 5 -1.0 -0.9137 0.0930 -1.096 -0.731
#> 6 -0.5 -0.4598 0.0894 -0.635 -0.285
#> 7 0.0 0.0358 0.0920 -0.144 0.216
#> 8 0.5 0.5732 0.0991 0.379 0.767
#> 9 1.0 1.1523 0.1101 0.936 1.368
#> 10 1.5 1.7731 0.1249 1.528 2.018
#> 11 2.0 2.4357 0.1442 2.153 2.718
#> 12 2.5 3.1401 0.1686 2.810 3.471
#> 13 3.0 3.8862 0.1989 3.496 4.276
#>
plot(fit.std)